Ultra-high throughput single-cell RNA sequencing by combinatorial fluidic indexing
Supplementary Website
scifi-RNA-seq overview
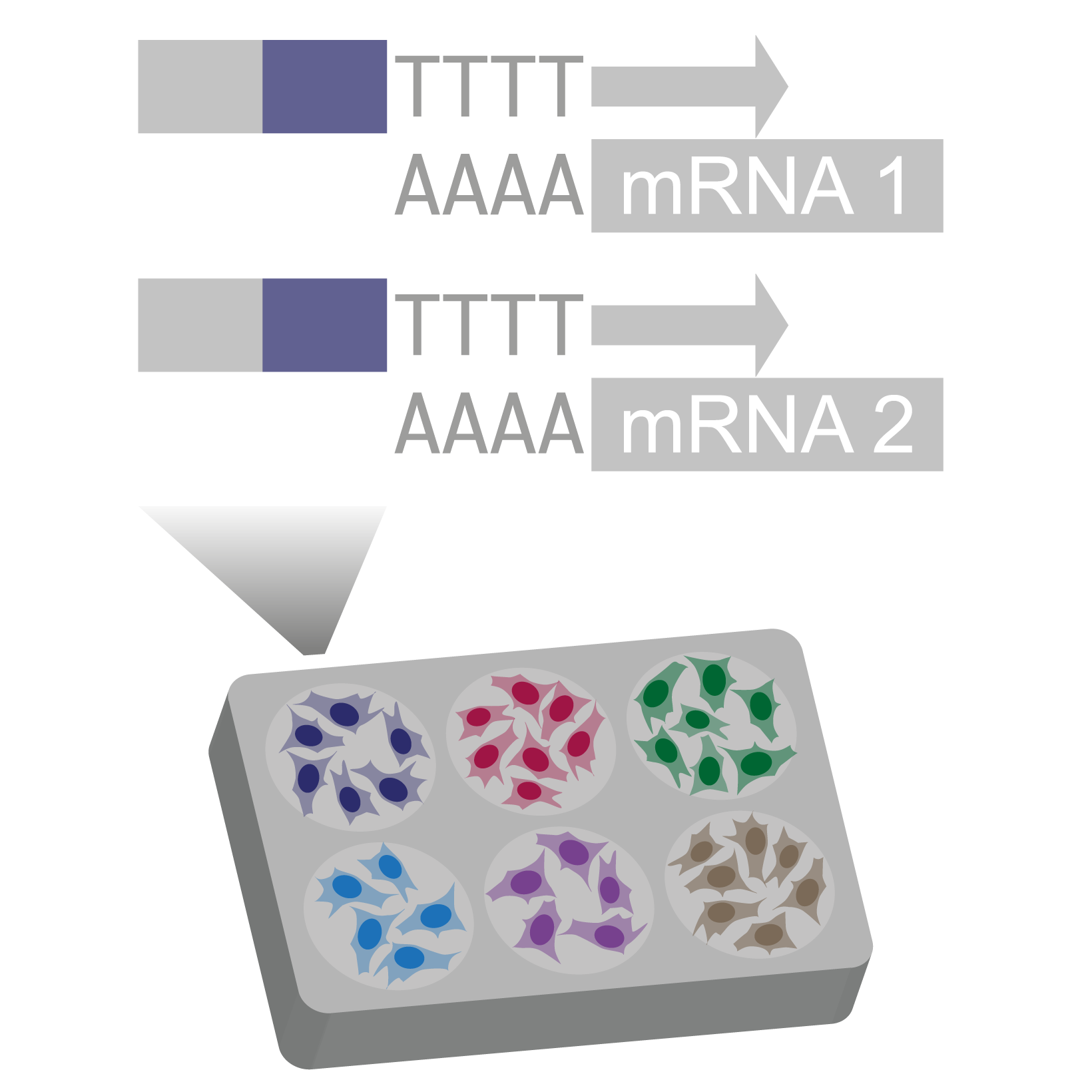
Pre-indexing single-cell transcriptomes
We use barcoded reverse transcription primers to index transcriptomes of permeabilized cells with a round1 barcode shared by all cells in the same well.
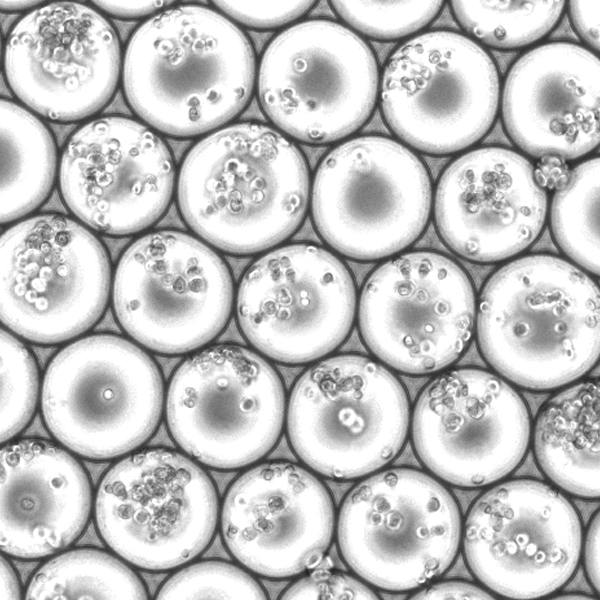
Microfluidics with droplet overloading
We randomly mix pre-indexed cells and overload the microfluidic droplet generator up to 100-fold. Multiple cells in the same droplet receive the same microfluidic round2 barcode.
Deconvolution of single-cell transcriptomes
Based on the combination of round1 and round2 barcodes, we obtain pure single-cell transcriptomes by deconvolution of sequencing reads.
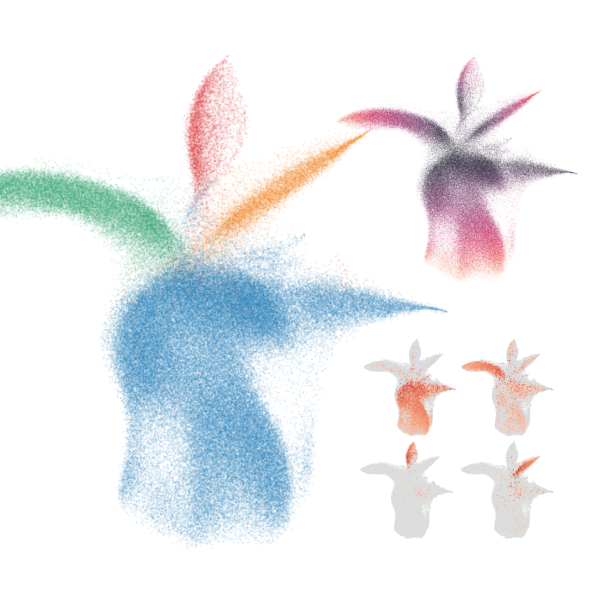
Ultra-high throughput scRNA-seq with sample multiplexing
scifi-RNA-seq enables ultra-high throughput scRNA-seq with multiplexing for thousands of samples, for instance for large perturbation screens at the single-cell level.
News and updates
| Date | Description |
|---|---|
| 2020/09/15 |
scifi-RNA-seq protocol designOur method is based on an innovative thermoligation step to connect round1 and round2 barcodes inside the emulsion droplets.

|
| 2019/12/18 |
scifi-RNA-seq pre-print published on bioRxivWe are excited to share our pre-print article on scifi-RNA-seq (DOI: 10.1101/2019.12.17.879304). |
Protocols and reagents
Updated protocols and oligonucleotide sequences for scifi-RNA-seq.
| Date | Description |
|---|---|
| 2020/09/15 |
scifi-RNA-seq step-by-step protocol (Version: 2021/01/25)Detailed step-by-step description of the scifi-RNA-seq method
The protocol contains a detailed listing of all required reagents and oligonucleotide sequences.
|
Data and software
Links to raw data archives and related resources.
| Name | Description and link |
|---|---|
| Raw and processed data | GEO accession: GSE168620. |
| Data processing pipeline | Git repository at Github: https://github.com/epigen/scifiRNA-seq |
| Code for scifi-RNA-seq paper | Git repository at Github: https://github.com/epigen/scifiRNA-seq_publication |
Citation
If you use this method in your research, please cite:
Paul Datlinger*, André F Rendeiro*, Thorina Boenke, Martin Senekowitsch, Thomas Krausgruber, Daniele Barreca, Christoph Bock. Ultra-high throughput single-cell RNA sequencing by combinatorial fluidic indexing. Nature Methods 18, 635–642 (2021). doi.org/10.1038/s41592-021-01153-z